In this laboratory, research is conducted to find appropriate intervention points for prevention and treatment of cancer and other health issues from the viewpoints of genomics and informatics. We are analyzing the dynamics of complex systems composed of many types of cells, such as cancer and inflammatory and immune diseases, by measuring large amounts of data at the genome level, and searching for specific phenomena that could serve as intervention targets and biomarkers for prevention and treatment, as well as their significance in disease. We are also working on the development of bioinformatics technology, including artificial intelligence, to extract and interpret essential information from large amounts of multi-dimensional genome sequences, histological images and other biological information by means of dimensional compression and visualization.

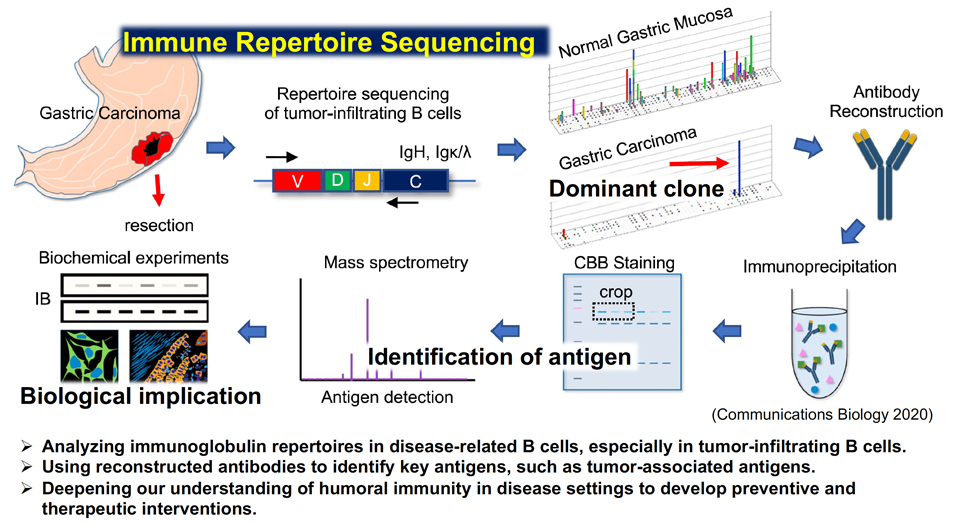
The antigen receptor genes of human lymphocytes acquire much enormous diversity through somatic recombination. The entire antigen receptor of a lymphocyte population is called an immune leptore, and advances in genome analysis technologies such as next-generation sequencers have made it possible to profile the immune leptore of individuals. The immune lepathore is thought to represent the individual's immune history due to external factors such as pathogens and food, and internal factors such as self-antigens and cancer antigens. We are developing a method to extract information important for health and hygiene, such as past diseases and lifestyle habits, as well as current disease status of individuals, by analyzing immune lepatores. In particular, in our immunorepatent study of gastric cancer and gastric mucosa tissue, we are using information analysis techniques such as machine learning and deep learning to clarify the overall picture of immune history against cancer antigens and H. pylori by integrating it with cancer genome analysis and microflora analysis, and to analyze information useful for cancer prevention and treatment. We are also attempting to synthesize antibodies from this immunorepatent data for the treatment and prevention of cancer.
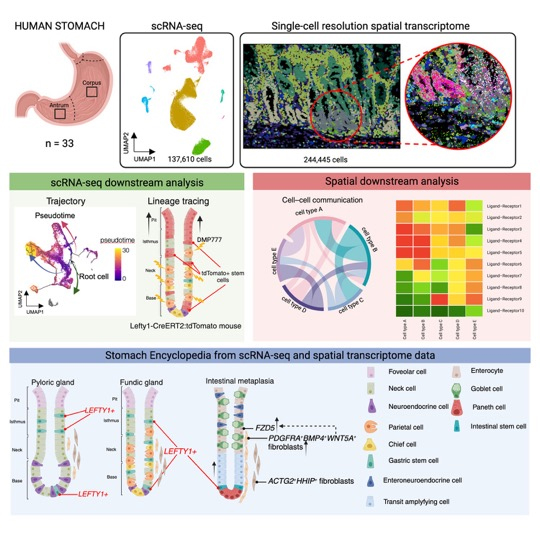
We are working on comprehensive understanding of cancer genomes using next-generation sequencing of human clinical cancer tissues. Comprehensive sequencing of cancer genomes provides information not only on driver genes that are direct therapeutic targets, but also on the genetic background that predisposes individuals to carcinogenesis and on environmental factors that contribute to carcinogenesis through mutation signature analysis. We are attempting to comprehensively capture this information and to find appropriate intervention points for prevention and treatment of cancers of public health importance in Japan through a combination of bioinformatics and experimental validation. Cancer genome sequencing of scirrhous gastric cancer has identified RHOA driver gene mutations and demonstrated that the mutation signature is different compared to that of normal gastric cancer, indicating that factors leading to carcinogenesis are different. Currently, we are working to comprehensively view the entire cancer tissue, including cancer cells, immune cells, and vascular stromal cells, from a genomics perspective, such as single cell analysis, to search for factors and intervention points for carcinogenesis and progression at a higher dimension.
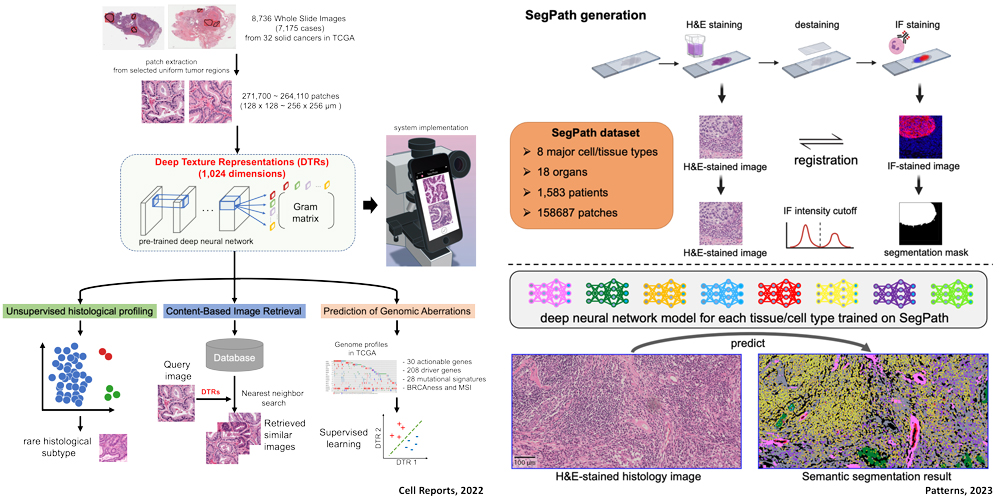
Extracting important information from large amounts of biological data is becoming an extremely important technology in a society inundated with large amounts of biomedical information. We are developing a method to extract essential cancer information from cancer histopathology images using deep learning technology, which will be useful for eliminating disparities in pathological diagnosis. We have also been working on the development of a new deep learning algorithm and have achieved results in international pathology image competitions with our original analysis method (Camelyon17). At present, through the integrated analysis of histopathological images and cancer genome information, we are developing a system that can directly lead from routine histological examinations that can be performed in small and medium-sized medical institutions to cancer genome medicine in core hospitals. We are also developing a system to search for similar histopathological images of cases with unknown diagnoses by structuring histopathological images of many cases into a database, and a technology to comprehensively detect different types of cells in histopathological images.