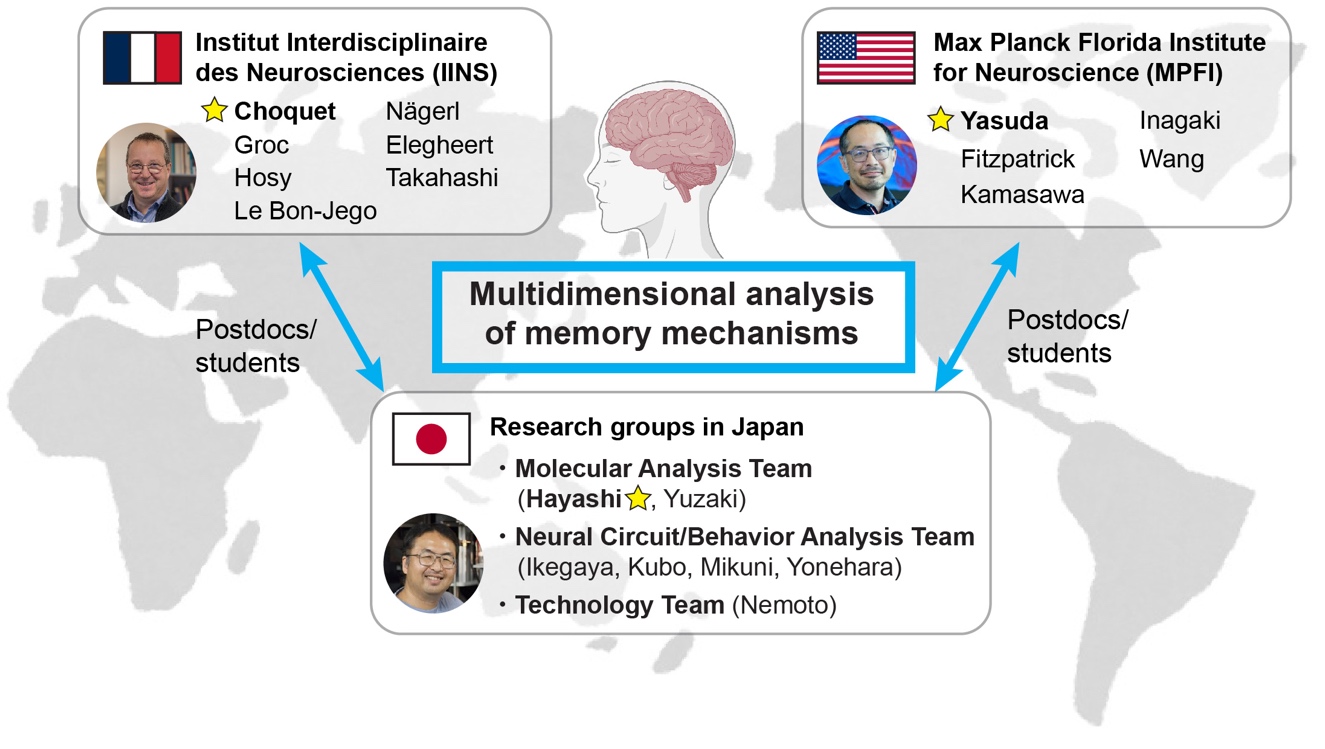
Research Organization
Molecular Analysis Team
Yasunori Hayashi (Kyoto University) Website
We are interested in the molecular mechanisms underlying synaptic plasticity. In particular, we are interested in how cellular signaling that leads to structural changes in the synapse is maintained in the long term beyond the induction of plasticity. To address this question, we use a combinatorial approach of biochemistry, two-photon imaging both in vitro and in vivo, FRET-FLIM, and mouse behavior.
Michisuke Yuzaki (Keio University) Website

By elucidating the molecular mechanisms underlying synapse formation and maintenance, our laboratory aims to develop specific synaptic connectors that will be useful in understanding the pathophysiology of neuropsychiatric disorders and in developing new therapeutics. We discovered that a new class of synaptic organizers, termed extracellular cellular scaffold proteins (ESPs), such as cerebellin (Cbln) and C1q-like family proteins, play critical roles in circuit- and activity-dependent synapse formation (Yuzaki, Annu Rev Physiol 2018). Recently, by combining structural elements from endogenous ESPs, we developed a synthetic synaptic organizer, CPTX, which restored synaptic function, motor coordination, spatial and contextual memory, and locomotion in mouse models of cerebellar ataxia, Alzheimer’s disease, and spinal cord injury. Our goal is to develop more synaptic connectors by uncovering new ESPs not only in the CNS, but also in the enteric and peripheral nervous systems. We welcome several graduate students and postdoctoral fellows to conduct research in our laboratory.
Neural Circuit and Behavior Analysis Team
Yuji Ikegaya (The University of Tokyo) Website

Our laboratory focuses on the plasticity in the microcircuitry of neuronal and glial cells, and our research topics cover fields ranging from development to disease. Our main targets are the limbic system, including the hippocampus and amygdala, and the cerebral cortex, where we search for mechanisms of cognitive functions such as memory and emotion, and functional alterations such as epilepsy and autism spectrum disorders. For these purposes, we use a variety of techniques such as electrophysiology, cell biology, functional imaging, and behavioral analysis, but we also routinely use machine learning for data analysis and neural information decoding.
Fumi Kubo (RIKEN Center for Brain Science) Website

Using zebrafish as a model, our lab studies the neural circuit mechanisms by which visual inputs produce goal-directed behavioral outputs. In particular, we aim to understand the roles of genetically defined neuron types and their circuit connectivity underlying the visually guided behaviors. In addition, we also study how visual functions are modified by past experiences. Taking advantage of the small and transparent zebrafish brain, our lab uses behavioral, genetic and optical techniques to analyze the visual circuits both at the cellular and network levels.
Takayasu Mikuni (Niigata University) Website

Our laboratory aims to understand the molecular and cellular mechanism of synaptic plasticity that underlies learning and memory. Towards this aim, we are developing new, original tools for monitoring synaptic activity, strength and molecular dynamics in hundreds to thousands of synapses in single neurons in the mouse brain in vivo. Using the tools combined with high-speed volumetric two-photon imaging, two-photon FRET-FLIM, two-photon photostimulation, and genome editing-based molecular labeling and manipulation techniques, we perform “synaptomic” analysis to comprehensively understand the single cell computation in the context of learning and memory.
Keisuke Yonehara (National Institute of Genetics) Website

We aim to understand the universal principles and diversity of sensory function emergence and its underlying structure by studying the visual system of mice and monkeys at multi-scales, including genes, molecules, cell types, circuits, neural processing, and behaviors. To this end, we combine various techniques such as genetics, two-photon imaging, electrophysiology, trans-synaptic labeling, single-cell transcriptomics, and machine learning.
Technology Development Team
Tomomi Nemoto (National Institute of Physiological Science) Website

We have advanced innovative bio-imaging methods by utilizing such advanced technologies as laser optics and nanomaterials. In particular, we facilitate our original world-leading ultra-deep, super-resolution, and ultra-high-speed imaging methods using nonlinear optical processes, including multiphoton excitation, for less-invasive observation and manipulation for living brain tissues and organs “in vivo.”
Our partners
US
Max Planck Florida Institute of Neuroscience
Website
France
Interdisciplinary Institute of Neuroscience – Bordeaux Neurocampus
Website

