Last Updated: 2015/07/21@@@
Softwares and data source by Yukinori Okada, M.D., Ph.D. [HomePage]
Softwares and data source used in Okada et al. Nature (2014)
Okada Y, Wu D, Trynka G et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature doi:10.1038/nature12873 [PubMed]
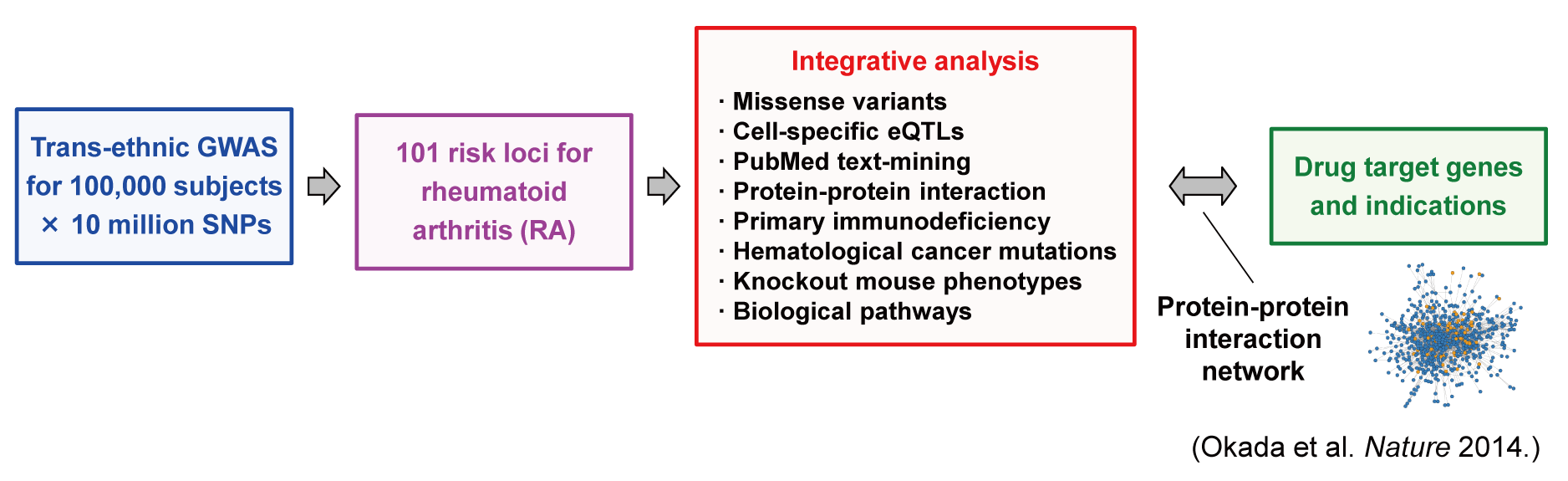
Summary statistics of RA GWAS meta-analysis
- Trans-ethnic RA GWAS meta-analysis (19,234 RA cases and 61,565 conrols) [Link]
- Eurpean RA GWAS meta-analysis (14,361 RA cases and 43,923 conrols) [Link]
- Asian RA GWAS meta-analysis (4,873 RA cases and 17,642 conrols) [Link]
- Summary results of RA risk SNPs in 101 risk loci [Link]
Softwares for 1KG imputation and GWAS meta-analysis
- Perl source codes for 1KG imputation reference panel preparation [Link]
- Perl source codes for splitting GWAS data / 1KG reference panel into chuncks [Link]
- Perl source codes for handling association analysis results [Link]
- Java package for genomic control (GC) correction [Link]
- Java package for GWAS meta-analysis [Link]
- R source codes for plotting Manhattan / QQ plots of GWAS P-values [Link]
Pleiotropy analysis using GWAS catalogue database
- Curated phenotype/SNP data from the GWAS catalogue (for data downloaded on January 31, 2013) [Link]
H3K4me3 histone mark enrichment analysis for GWAS signals
- Software for enrichment analysis and histone mark data (from Raychaudhuri lab and the Broad Instiute) [Link]
Softwares and data source for in-silico pipeline to prioritize biological genes from GWAS risk loci
- Java package for calculating LD between SNPs and 1KG reference data [Link]
- R source codes for assigning LD regions to SNPs [Link]
- Perl source codes for assiging UCSC genes into LD regions [Link]
- Functional annotation of SNPs (Annovar software) [Link]
- eQTL data for PBMC (from http://genenetwork.nl) [Link]
- Cell-specific eQTL data (from ImmVar project) [Link]
- PubMed text-mining (GRAIL software) [Link]
- Protein-protein interaction analysis (DAPPLE software) [Link]
- Primary immunodificiency (PID) gene list (from Journal web site) [Link]
- Cancer somatic mutation gene list (from Journal web site) [Link]
- Knockout mouse phenotype and gene list (from Journal web site) [Link]
- Molecular pathway analysis (MAGENTA software) [Link]
Softwares and data source for GWAS-drug target overlap analysis
- Drug target gene list (from Journal web site) [Link]
- Perl source codes for overlap enrichment analysis [Link]
Softwares used in Okada et al. ASHG Meeting (2009)
Okada Y, Yamamoto K, Yamada R. Estimation of the Null Distribution of Fisherfs Exact Test P-values in GWA Studies. ASHG Meeting (2009)
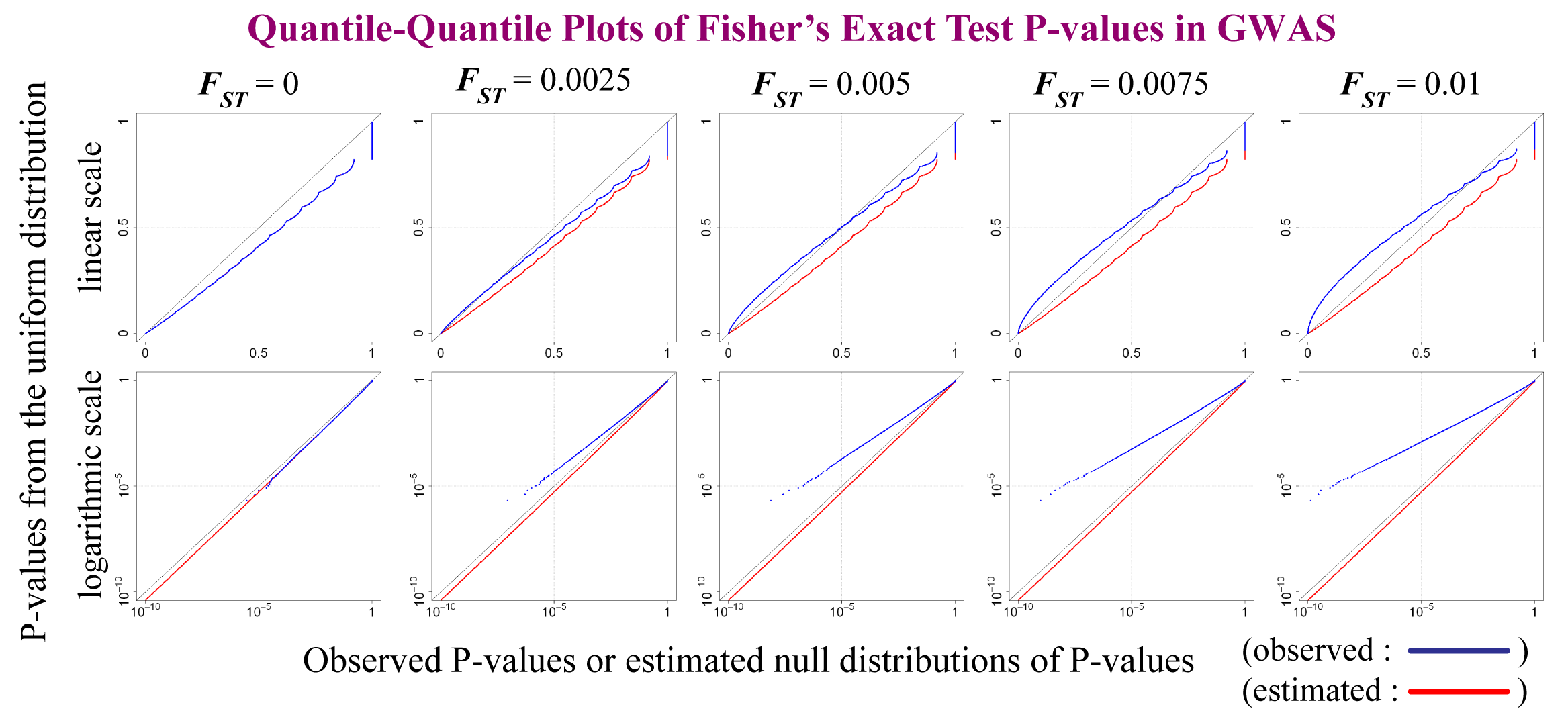
Estimation of null distribution of Fisher's exact test P-values for 2x2 SNP tables in GWAS [Poster]
- Java package for null distribution estimation [Link]
Corresponding to : Yukinori Okada, M.D., Ph.D.
Web site : http://plaza.umin.ac.jp/~yokada/ [HomePage]
E-mail : yokada (at) src.riken.jp, yokada-tky (at) umin.ac.jp

